Master transcription-factor binding sites constitute the core of early replication control elements
San Diego BioMed congratulates the Gilbert Lab for their recent publication featured in The EMBO Journal!
Learn and read the full manuscript @ https://www.embopress.org/doi/full/10.1038/s44318-025-00501-5
Professor Gilbert provided an engaging commentary on this manuscript and his lab’s work below –
Professor Gilbert’s key takeaway: “Timing is everything.”
“DNA has become a household term to define the ‘being’ of any living thing and is even clumsily used to refer to inanimate objects. Most of us are familiar with DNA’s 4 letter alphabet, which encodes the blueprint for life. Today, we can chemically synthesize any DNA code, we can create billions of copies of that code in a test tube, and we can even cure diseases by correcting mistakes in the code. It must come as quite a surprise then, that we still do not know the genetic code that tells cells how to replicate DNA. This is a major mystery in molecular biology and is the motivation for research in the Gilbert lab at San Diego BioMed.
The Gilbert lab believes that the secret to understanding how DNA replicates lies in understanding why it bothers to replicate the code in any particular order; after all, it is commonly thought that the sole purpose of DNA replication is to make one complete copy of the DNA before the cell divides into two. The Gilbert lab likes to remind people that, in a living cell, it’s not just DNA that replicates. Rather, the entire structure of chromosomes, including all of their associated components, must be dismantled, the DNA copied, and all the infrastructure rebuilt. This rebuilding process imparts flexibility to change the infrastructure and Mother Nature is too resourceful to squander this opportunity.
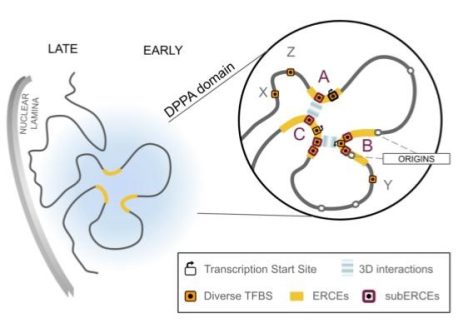
The Gilbert lab has spent over 30 years compiling evidence that chromosomes are replicated in segments or “domains” and that the temporal order in which domains are replicated dictates what type of chromosomal infrastructure they assemble. Indeed, the Gilbert lab has shown that different types of cells replicate chromosome domains in a different order and cells from diseased tissues have unique alterations in this replication timing program. In 2019, by systematically removing parts of chromosomes, the Gilbert lab discovered specific control centers that determine this temporal order. However, they were not able to sufficiently pinpoint where these control centers are in order to understand how they work.
In the current work, published by The European Molecular Biology Organization (EMBO J.), the Gilbert lab researchers–Dave Gilbert, Takayo Sasaki, and Satoshi Uchino–have shown that these control centers contain numerous binding sites in the DNA for multiple proteins known as “master transcription factors”, whose job is to turn individual genes on and off to alter the identity of cells. This work unveils a simple but elegant means by which cells coordinate changes in gene regulation with changes in replication timing. And the purpose is quite sensible; it is known that turning genes on and off can be a fleeting event that fades away as cells experiment with new fates and revert to their old ways. But, if the master transcription factors are diverse and abundant enough to bind all the sites in the replication control centers, they can trigger changes in replication timing that could alter the infrastructure of chromosomes and make the switch to the new cell type irreversible, whether it be a normal or diseased cell type. What is still not known is whether these switches are encoded in the alphabet of the DNA and what that code might be. Understanding that code could allow us to alter the fates of cells or switch diseased cells back to normal. Stay tuned for the Gilbert lab’s subsequent studies!”


